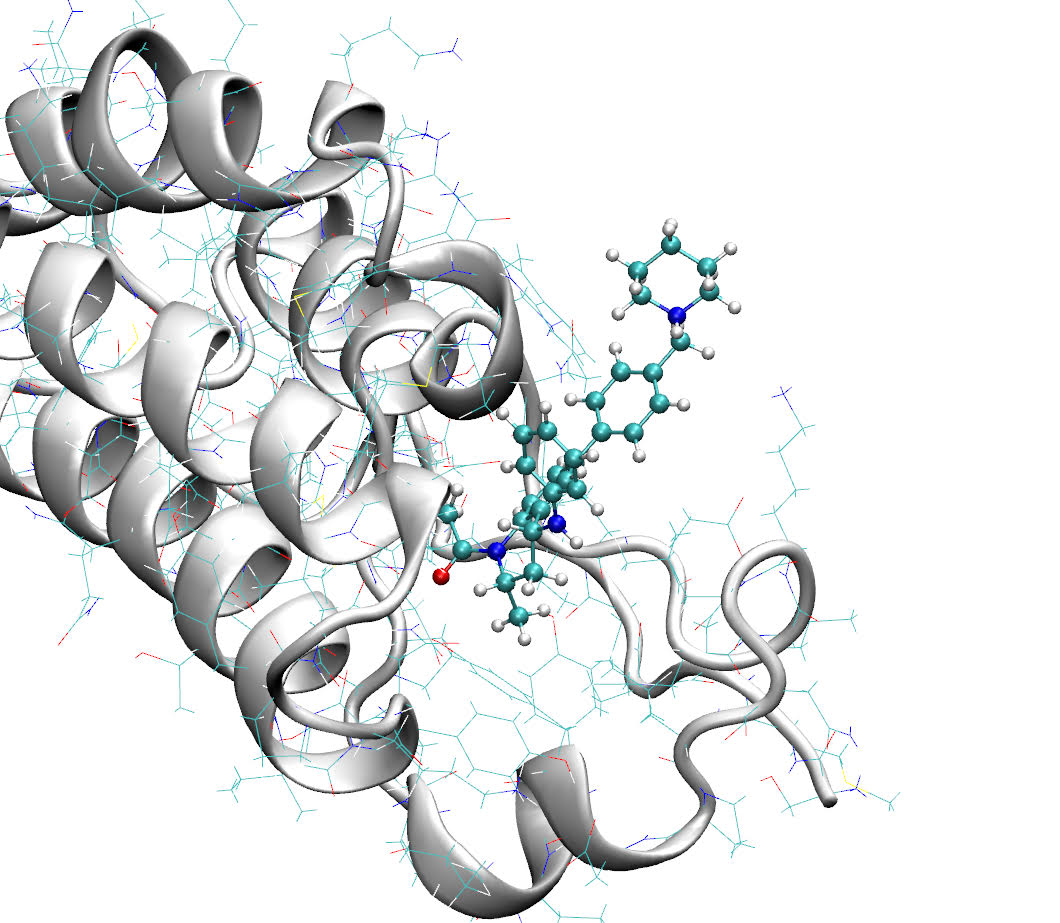
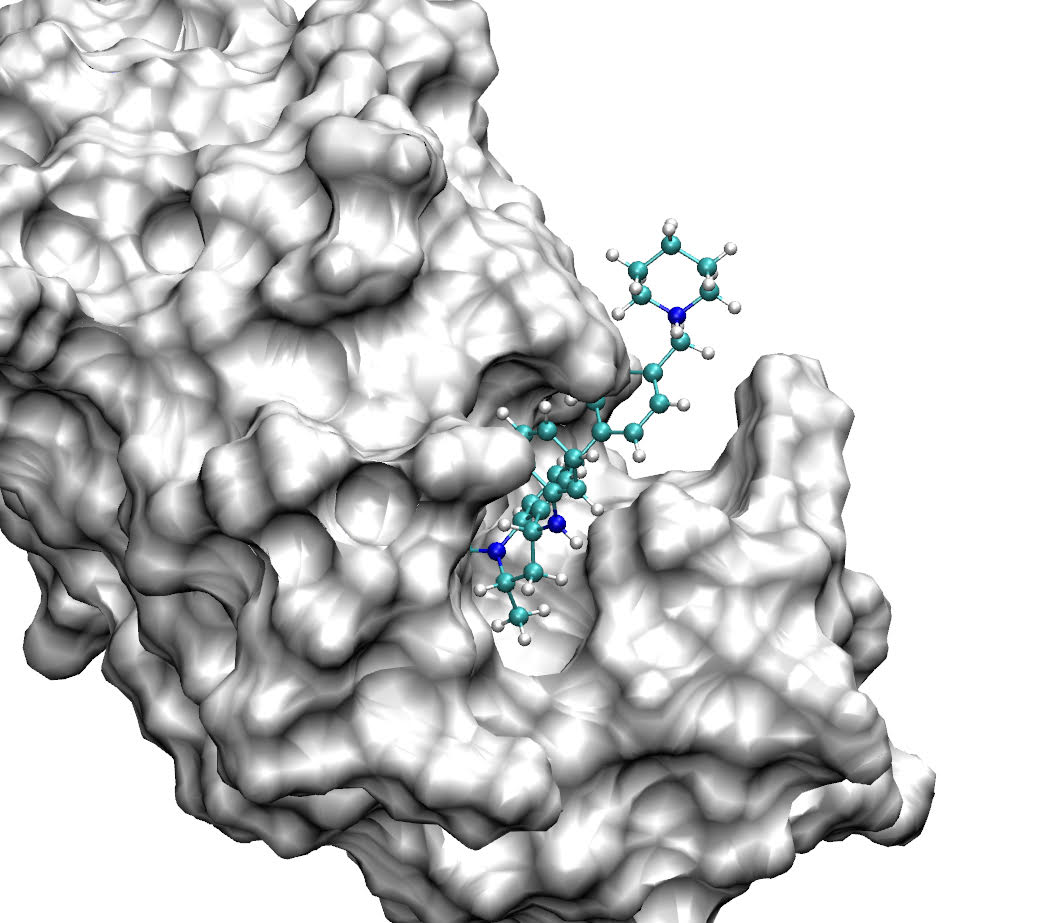
A joint UCL / Janssen team has performed a statistically robust head-to-head comparison between the Centre for Computational Science’s high performance computational method, TIES, and that offered by a commercial provider, Schrödinger Inc, FEP+, for calculating relative free energies of binding of candidate drugs to target proteins. TIES and FEP+ use different so-called “alchemical” free energy methods. A robust ensemble-based protocol is applied for the evaluation of the computational results and their associated errors. In the study, TIES produced the most reliable results. More compute-intensive “replica-exchange” methods, hard-wired within FEP+ but available optionally within TIES, manifest systematic underestimations of these free energy differences; in particular, FEP+ predictions degrade substantially as simulation times are extended.
S. Wan, G. Tresadern, L. Perez-Benito, H. van Vlijmen, P. V. Coveney, “Accuracy and Precision of Alchemical Relative Free Energy Predictions With and Without Replica-Exchange”, Advanced Theory and Simulations, in Press (2019)
DOI: 10.1002/adts.201900195
You can access the paper here.
Publication date: 27 November, 2019
