SARS-CoV2 has rapidly spread worldwide since December 2019, and early modelling work of this pandemic has assisted in identifying effective government interventions. The UK government relied in part on the CovidSim model developed by Professor Neil Ferguson’s team at the MRC Centre for Global Infectious Disease Analysis at Imperial College London, to model various non-pharmaceutical intervention strategies, and guide its government policy in seeking to deal with the rapid spread of the COVID-19 pandemic during March and April 2020.
CovidSim, albeit a sophisticated model, contains a large degree of uncertainty in its predictions, due to its inherent nature. The model is subject to different sources of uncertainty, namely parametric uncertainty in the inputs, model structure uncertainty (i.e. missing epidemiological processes) and scenario uncertainty, which relates to uncertainty in the set of conditions under which the model is applied. A result of these different types of uncertainty combined was the serious underestimation of the first wave: predictions of COVID-19 deaths in the influential Report 9 issued in March were about half the number of deaths that actually occurred in the UK, when simulating the most comparable scenarios -albeit with some differences, such as lockdown occurring later than simulated.
Scientists from VECMA and CompBioMed analyzed CovidSim by undertaking an extensive parametric sensitivity analysis and uncertainty quantification of the publicly available code. From the over 900 parameters that are provided as input to CovidSim, a key subset of 19 was identified to which the code output is most sensitive. It was found that the uncertainty in the code is substantial, in the sense that imperfect knowledge in these inputs will be magnified to the outputs, up to the extent of ca. 300%. Most of this uncertainty can be traced back to the sensitivity of 3 parameters. Compounding this, the model can display significant bias with respect to observed data, such that the output variance does not capture this validation data with high probability. The large variance in the prediction of an outcome (coronavirus deaths, in this case) can be seen through the example of the following figure, reported in the study:
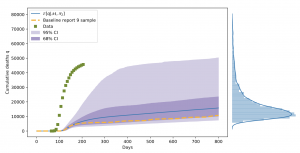
Figure: The mean cumulative death prediction for a calculated epidemic scenario, plus confidence intervals, and at the right, the pdf of the total death count after 800 days. It can be seen that the predicted cumulative deaths may vary by as much as 100% (i.e. double), while still remaining within one standard deviation in terms of confidence. (Results were obtained using a computational budget of 3000 CovidSim evaluations per scenario. Day 0 corresponds to January 1st, 2020. Superimposed (green squares) are the observed cumulative death count data for the UK obtained from gov.uk. The first data point falls on 6th March 2020, corresponding to day 66. The striped line is a single sample from CovidSim.)
The VECMA and CompBioMed teams concluded that quantifying the parametric input uncertainty is not sufficient, and that the effect of model structure and scenario uncertainty cannot be ignored when validating the model in a probabilistic sense. What is more, the scientific teams of the two EU projects call for a better public understanding of the inherent uncertainty of models predicting Covid-19 mortality rates, saying they should be regarded as “probabilistic” rather than being relied upon to produce a particular and specific outcome. They maintain that future forecasts used to inform government policy should provide the range of possible outcomes in terms of probabilities to provide a more realistic picture of the pandemic framed in terms of uncertainties.
VECMA and CompBioMed Principal Investigator Professor Peter Coveney noted: “There is a large degree of uncertainty in the modelling used to guide governments’ responses to the pandemic and this is necessary for decision makers to understand.
“This is not a reason to disregard modelling. It is important that these simulations are understood in terms of providing a range of probabilities for different outcomes, rather than a single fixed prediction of Covid-19 mortality.”
“Because of this uncertainty, future forecasts of the death rates of Covid-19 should be based not on an individual simulation, but on lots of different simulations of a code, each with slightly adjusted assumptions. Predictions based on this method, though still highly uncertain, will provide a more realistic picture of the pandemic.”
Professor Coveney added: “Our findings are important for government and healthcare policy decision making, given that CovidSim and other such epidemiological models are – quite rightly – still used in forecasting the spread of COVID-19. Like predicting the weather, forecasting a pandemic carries a high degree of uncertainty and this needs to be recognised.
“Finally, our modelling has only been possible because Professor Neil Ferguson’s team open sourced their code. Not all models being used in Government briefings are open in that way. We urge other research groups to follow Imperial’s lead and adopt an open science approach.”
The study was undertaken on behalf of the Royal Society’s RAMP initiative and is now available as a publication preprint. The work was funded as part of the European Union Horizon 2020 research and innovation programme under grant agreements #800925 (VECMA project) and #823712 (CompBioMed2 Centre of Excellence), as well as the UK EPSRC for the UK High-End Computing Consortium (EP/R029598/1).
